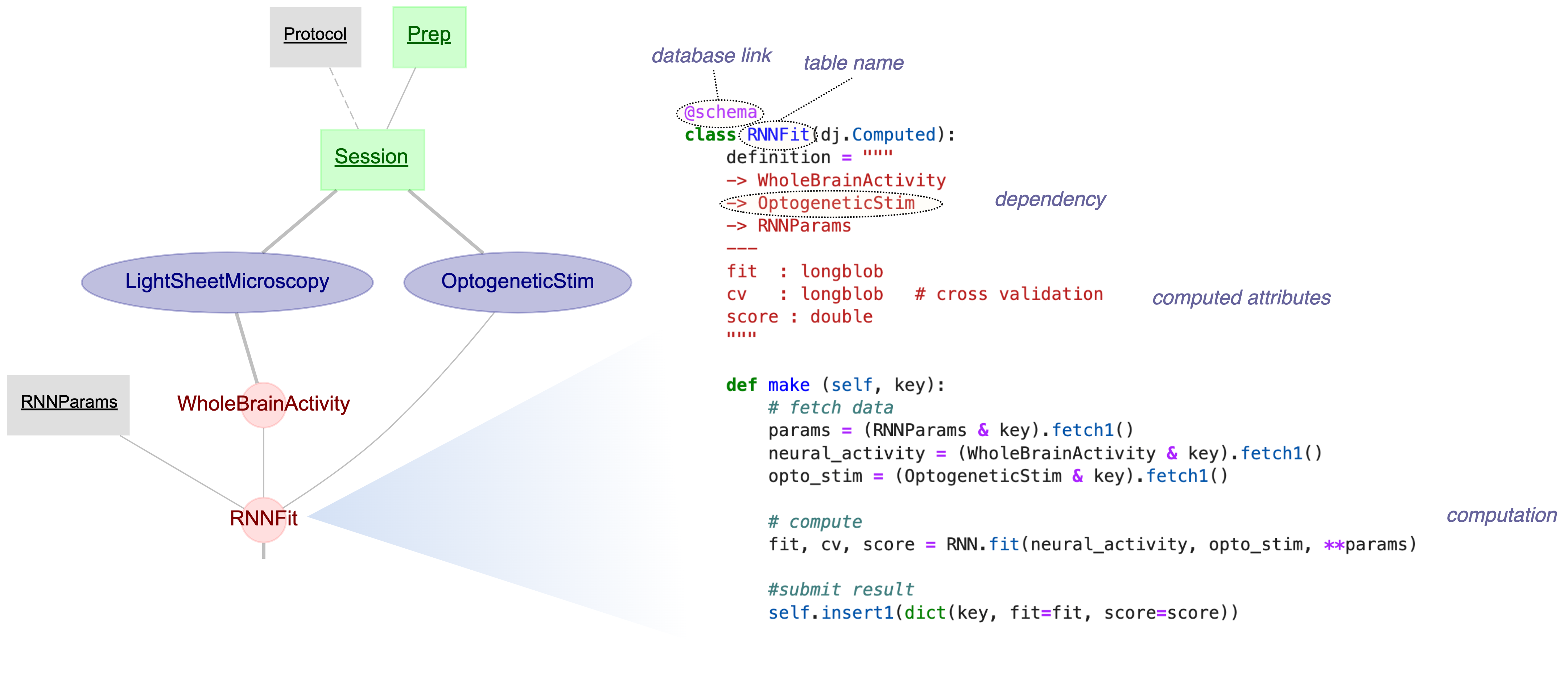
DataJoint is a framework for scientific data pipelines based on the Relational Workflow Model — a paradigm where your database schema is an executable specification of your workflow.
- Tables represent workflow steps — Each table is a step in your pipeline
- Foreign keys encode dependencies — Parent tables must be populated before child tables
- Computations are declarative — Define what to compute; DataJoint handles when
- Results are immutable — Full provenance and reproducibility
Documentation: https://docs.datajoint.com
📘 Upgrading from legacy DataJoint (pre-2.0)? See the Migration Guide for a step-by-step upgrade path.
| PyPI |

|
Conda |

|
Tests |
|
| License |

|
Citation |

|
Coverage |
|
pip install datajointor with Conda:
conda install -c conda-forge datajoint- Documentation — Complete guides and reference
- Tutorials — Learn by example
- How-To Guides — Task-oriented guides
- API Reference — Complete API documentation
- Migration Guide — Upgrade from legacy versions
- DataJoint Elements — Example pipelines for neuroscience
- GitHub Discussions — Community support
See CONTRIBUTING.md for development setup and guidelines.